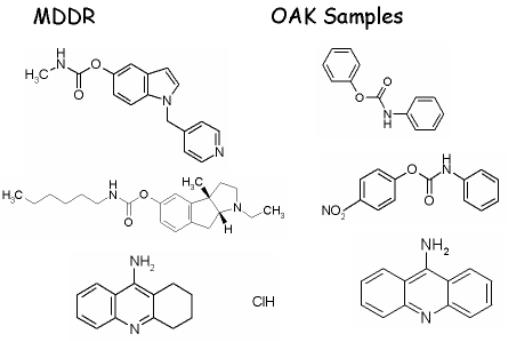
How to select promising candidates from supplier databases!
|
Problem: Find potential acetylcholinesterase inhibitors (AChEI) in a commercial screening compound database.
|
|
Strategy:Develop a profile for acetylcholinesterase inhibitors by looking for other activities that are important for this class of compounds.Overlap in multidimensional activity space predicted compounds and compounds with experimentally known activities. |
Hypothesis:Compounds that are in the same region in the activity space have a very high probability of having similar biological activities.
|
Experiment (1):We select all acetylcholinesterase inhibitors from the principal list in MDDR. Principal list means only those compounds are selected that are the principal compounds in a patent or report.We predict the PASS parameters with pa>80% for all these compounds. We find a limited number (18) of other activities that are important for some of these compounds. We use the set of 18 activities to screen in silico the OAK Samples database (OAK).
|
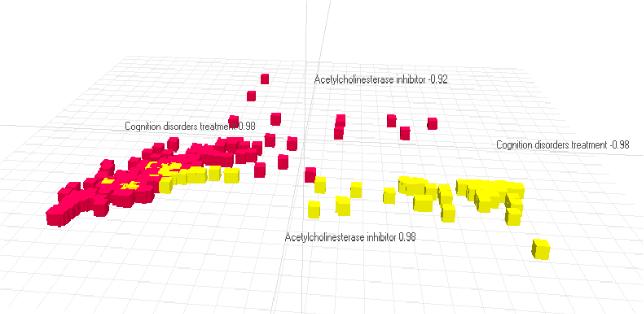
We combine the compounds from MDDR and all compounds with a PA>50% from OAK in an Excel spreadsheet. (If you want, you can download (0.8 MB) the zipped spreadsheet following this link. The spreadsheet contains the structures and was produced with ISIS for EXCEL.) We display the data using miner3d.excel, plotting first the activities "acetylcholinesterase inhibitor" against "cognition disorder"
|
 |
|
| and we see a nice clustering of the red (MDDR) and the yellow (OAK) points | |
| The next pictures shows the points plotted in three dimensions, "acetylcholinesterase inhibitor", "cognition disorder" and "cholinergic agonist". |  |
| The final picture shows as fourth activity "miotic". The points were filtered so that more points with similar values for all activities become visible (balls). |  |
| The following table shows a few selected compounds, displayed as balls in the previous plot. | |
|
|
|
Experimemt (2):The same as in experiment (1), but we use a set of activities that might be relevant for actylcholinesterase inhibitors. (We thank Prof. U. Jordis from the Techncal University in Vienna for his suggestions.) We used in addition to MDDR and OAK Samples, a set of compounds from the NCI database that had pa > 0.7. Download the EXCEL spreadsheet with the calculated PASS parameters, and try for yourself analysing the data using miner3d.excel |
We created the plot AChE vs. cognition disorders treatments vs cholinergic agonist from Experiment (1) and the plot AChE vs. ACh nicotinic agonist vs. ACh nicotinic antagonist from Experiment (2) structures. form both plots, we select a cluster of compounds that overlapped with the structures of MDDR. This is fairly arbitrary way of selecting compounds, and in spite of this we found a large overlap among the selected compounds. This encourages us that or hypothesis, see above works. You can download here the EXCEL spreadsheet with the results. |
|
Plotting the activities:
|
You need the Chime add-in to see the structures |
|
|
|
Experimemt (3):An experienced chemist looking at the results of experiment 2, the compounds that are selected as potential AChE inhibitors, recognizes that these substances are not very drug like. Most of the compounds are flat, missing chiral centers and are polyaromatic compounds. Instead of using the limited OAK Samples database, we could use ACD-SC. This is a database of over 1.5 million compounds containing structures of most of the suppliers offering screening compounds. This database is very large for desktop machines and was not installed on our system. The chance of finding drug like compounds would have been much larger. |
However, we can try to find a real candidate using a database like MDDR that contains only drug like compounds. We searched for "Acetylcholinesterase Inhibitor" in the principal list of 27554 compounds and found 141 hits. |
|
We subtracted these from the principal list, exported the SDFile, generated the PASS parameters, and selected all compounds with Pa-Pi > 0.3. We generated a spreadsheet containing these compounds, the 141 experimentally known AChE inhibitors and as comparison a list of compounds from the commercially available Maybridge database. You can download the zipped file mddr_pricipal-ache_maybridge.zip. We looked for compounds who overlapped with the known AChE inhibitors in the biological space consisting of AChE inhibitor, cognition disorder treatment, antiparkinsonian and cholinergic agonist. |
We selected a few compounds and looked at them again in MDDR. We found one compound that was not classified as AChE inhibitor in the "Activity" field of MDDR, but was tested active as AChE inhibitor, according to the description in the "Biology" field. |
|
This is the text in the section Biological Activity in MDDR: ACTION - Neuroprotectant and cerebral function improving agent, acetylcholinesterase inhibitor (IC50=3.7 mcM); compound may be useful in the treatment of Alzheimer's disease and other memory disorders. Other compounds from a series of aminonaphthalene derivatives are the |
following:183158 183159 183160 183161 183162 183163.
We believe this is a very convincing result showing the ability of PASS and miner3d.excel to be helpful in identifying new promising AChE inhibitors. |
Biological DiversityIn the above experiment, we also included compounds from the Maybridge database with Pa-Pi > 0.5. None of the compounds overlapped with known AChE inhibitors in the biological space consisting of AChE inhibitor (AChEI) and cognition disorder (CogD) treatment. However, we found another application of using the biological spaces. We only looked at the Maybridge compounds and plotted these data in other biological spaces. |
We found some clustering using a 5 dimensional space using ACheI, CogD, cholinergic agonist, antiparkinsonian, and adenosine A1 receptor agonist. A chemist selecting screening compounds would select a few representatives from each cluster. In close comparison to the use of chemical diversity when selecting screening samples, a chemist can select now compounds using "biological diversity". |